Cancer is a complex and heterogeneous disease that poses a major challenge for effective treatment. One of the most promising strategies to harness the power of the immune system to fight cancer is to target neoantigens, which are novel peptides generated by somatic mutations in tumor cells. Neoantigens can elicit specific and potent immune responses by T cells, which can recognize and eliminate cancer cells. However, identifying neoantigens that are relevant for immunotherapy is not a trivial task, as it requires analyzing large amounts of genomic and proteomic data from tumor and normal tissues, as well as predicting the binding affinity and immunogenicity of thousands of candidate peptides. This is where artificial intelligence (AI) can play a crucial role, by providing computational tools and algorithms that can accelerate and improve the accuracy of neoantigen identification and prioritization.
Getting Data for Neoantigen Identification
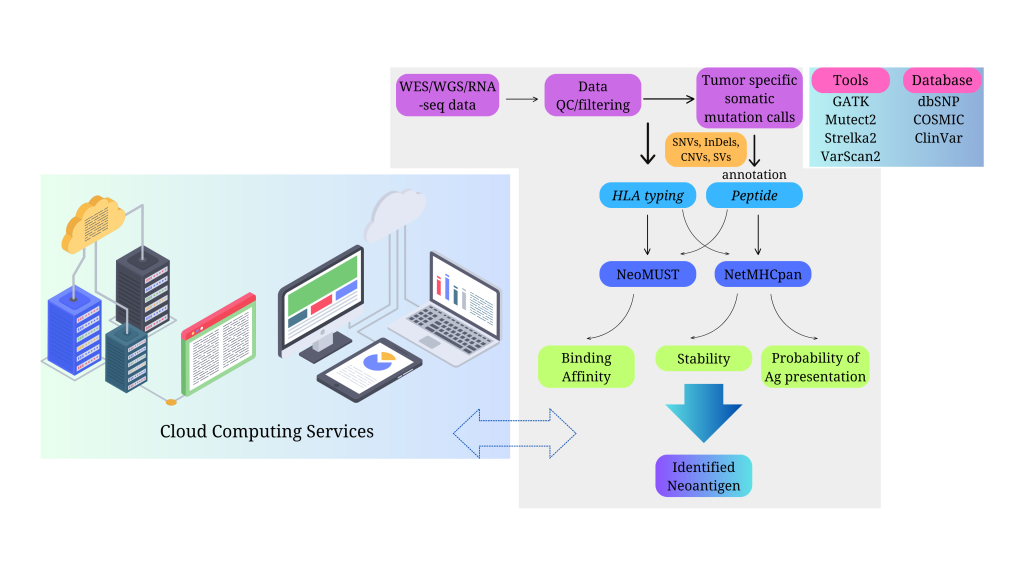
The first step in neoantigen identification is to obtain high-quality sequencing data from tumor and normal samples, preferably from the same patient. This can be done using next-generation sequencing (NGS) technologies, such as whole-exome sequencing (WES), whole-genome sequencing (WGS), or RNA sequencing (RNA-seq). These methods can generate millions of reads that cover the coding and non-coding regions of the genome and transcriptome, respectively. The sequencing data can then be aligned to a reference genome and annotated with gene and protein information.
The next step is to identify somatic mutations that are present in the tumor but not in the normal tissue, such as single nucleotide variants (SNVs), insertions and deletions (indels), copy number variations (CNVs), or structural variations (SVs). These mutations can be detected using various bioinformatics tools, such as Mutect2, VarScan2, or GATK. The identified mutations can then be filtered based on their frequency, quality, and functional impact, using databases such as dbSNP, COSMIC, or ClinVar.
The final step is to translate the somatic mutations into candidate neoantigens, which are peptides of a certain length (usually 8-11 amino acids for MHC class I and 15-25 amino acids for MHC class II) that contain the mutated residue. This can be done using tools such as pVACtools, MuPeXI, or NetMHCpan. The candidate neoantigens can then be ranked based on their predicted binding affinity to the patient-specific MHC molecules, which are encoded by the highly polymorphic HLA genes. The HLA typing of the patient can be inferred from the sequencing data using tools such as OptiType, HLAminer, or POLYSOLVER.
AI Algorithms for Neoantigen Prediction
The conventional methods for neoantigen prediction rely on empirical rules or statistical models that are based on limited experimental data and biological knowledge. However, these methods have several limitations, such as low sensitivity, specificity, and reproducibility, as well as high computational cost and complexity. Moreover, these methods do not account for the immunogenicity of the candidate neoantigens, which is the ability to induce an immune response by T cells.
AI algorithms, especially machine learning (ML) and deep learning (DL) techniques, have emerged as powerful alternatives to overcome these challenges. ML and DL algorithms can learn from large amounts of data and extract complex patterns and features that are relevant for neoantigen prediction. ML and DL algorithms can also integrate multiple types of data, such as genomic, proteomic, transcriptomic, epigenetic, or immunological data, to provide a more comprehensive and accurate picture of the neoantigen landscape.
Some examples of AI algorithms that have been applied to neoantigen prediction are:
• pMTnet, a DL-based algorithm that can predict the binding between neoantigens, MHC molecules, and T cell receptors (TCRs) using a multi-task learning framework. pMTnet outperformed conventional methods in terms of accuracy and efficiency.
• PELEUS, an ML-based algorithm that can rank neoantigens based on their immunogenicity using a novel scoring system that incorporates multiple features, such as peptide-MHC stability, TCR diversity, tumor expression level, and clonality. PELEUS showed superior performance in identifying clinically relevant neoantigens.
• TumorMap, an ML-based algorithm that can visualize and cluster neoantigens based on their similarity and relevance for immunotherapy using a self-organizing map approach. TumorMap can facilitate the selection of neoantigens for personalized cancer vaccines or TCR-T cell therapy.
TCR-T Cell Therapy for Neoantigen Targeting
One of the potential applications of neoantigen identification is to design TCR-T cell therapy, which is a type of cancer immunotherapy that uses genetically engineered T cells that express a specific TCR that can recognize a neoantigen. TCR-T cell therapy has several advantages over other types of immunotherapy, such as:
• TCR-T cells can target neoantigens that are derived from intracellular proteins, which are more abundant and diverse than surface antigens that are targeted by CAR-T cells.
• TCR-T cells can recognize neoantigens in the context of both MHC class I and II molecules, which can enhance the activation and proliferation of both CD8+ and CD4+ T cells.
• TCR-T cells can induce a more durable and memory-like immune response, which can prevent tumor relapse and resistance.
However, TCR-T cell therapy also faces some challenges, such as:
• The identification and isolation of neoantigen-specific TCRs is difficult and time-consuming, as it requires screening a large pool of T cells from tumor or peripheral blood samples.
• The affinity and specificity of the TCRs need to be optimized to ensure effective tumor recognition and avoid cross-reactivity with normal tissues.
• The safety and efficacy of the TCR-T cells need to be evaluated in preclinical and clinical trials, which can be costly and lengthy.
To overcome these challenges, AI algorithms can play a key role in facilitating the development of TCR-T cell therapy, by:
• Predicting the binding affinity and immunogenicity of neoantigens and their corresponding TCRs, which can reduce the experimental workload and increase the accuracy of neoantigen selection.
• Designing novel or enhanced TCRs that have higher affinity, specificity, and stability, which can improve the therapeutic potential and safety of TCR-T cells.
• Analyzing the clinical data and outcomes of TCR-T cell therapy, which can provide insights into the mechanisms of action, biomarkers of response, and factors of resistance.
Conclusion
AI assisted neoantigen identification is a promising approach for personalized cancer immunotherapy, especially for TCR-T cell therapy. AI algorithms can provide powerful tools and solutions for neoantigen prediction, prioritization, delivery, and validation. However, there are still some limitations and challenges that need to be addressed, such as the availability and quality of data, the interpretability and robustness of algorithms, the integration and standardization of workflows, and the ethical and regulatory issues. Therefore, more research and collaboration are needed to advance the field of AI assisted neoantigen identification and to translate it into clinical practice.