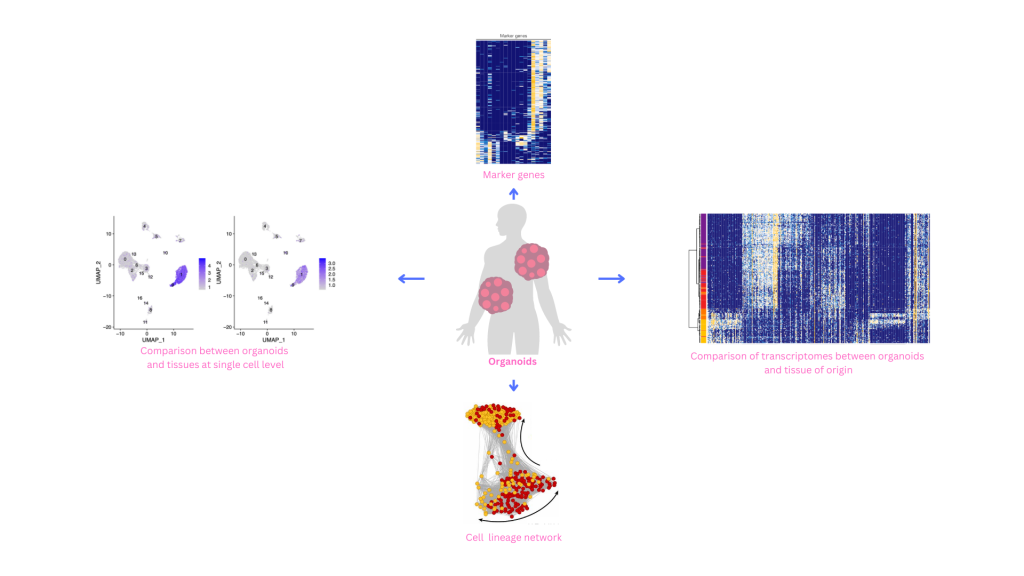
Organoids have served an efficient tool for drug screen, disease study, and personalized medicine. Characterization and evaluation of between the organdies and their origin of tissue remain critical for validation of the data obtained. Single cell sequencing-based technology provides a high-throughput method for testing and characterizing the organdies.
New high-throughput single-cell transcriptomic methods based on droplet microfluidics, combinatorial barcoding, or microwell technologies have opened up new path to quantitative comparisons of cell states, identities, and lineage. These high-throughput methods are enabling the analysis of human 3D organoids across a range of environmental, genetic, and temporal scales. High-throughput methods will make it possible to quantitatively assess cell states between organoids, batches, protocols, and various environmental perturbations; between replicates, clones, individuals from the same or different populations, and patients with genetic alterations; and each of the genetic or environmental impacts on cell state can be assessed over a time course.
Single-cell transcriptomics further allows the analysis of fate specification — even when cells are not correctly arranged — and has become the method of choice for protocol optimization and the analysis of variability, lineages, response to morphogens or toxic chemicals, and disease-relevant mutations. The technology is based on the key assumption that the subset of genes actively transcribed by a cell is a reliable readout for cell types and that the establishment of this activity pattern follows a trajectory recapitulating cell fate specification.Therefore, heat map visualization of cells ordered according to their expression profiles along the lineage can reveal a temporal sequence of gene regulatory events during differentiation.